Sample Guidelines scRNA
- General Cell
requirements - Sample preservation
and thawing - Nuclei Isolation
- FLEX
- ATAC/ multiome
- Power calculation tools
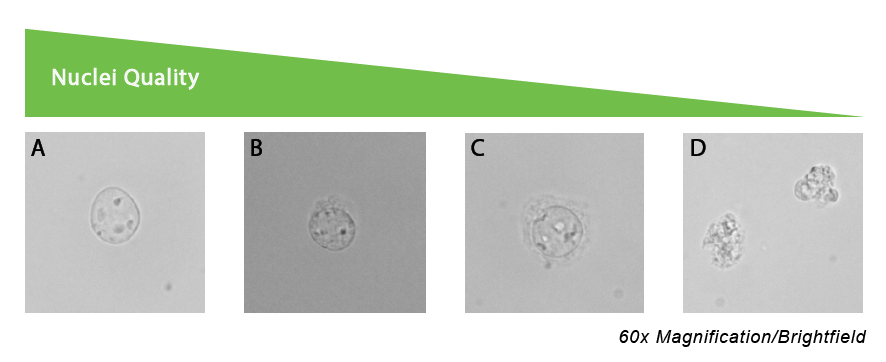
Cell cryopreservation is supported by both 10X and BD Rhapsody technologies. Protocols may vary depending on the cell type. Here is a general guideline for cell preparation for single cell protocols. We recommend that you use a test sample to verify that this protocol is suitable for your samples and to check viability after thawing, as cryopreservation can have a major impact on cell viability.
Thawing: One or more protocols may be applicable for a specific sample type and the most appropriate protocol should be used for thawing single cell suspensions for use with 10x Genomics Single Cell assays. This handbook describes different protocols depending on the sample type.